 3D Reconstruction of Human Olfactory System
3D Reconstruction of Human Olfactory System
Authors: Victoria F Low, Chinchien Lin, Shan Su, Mahyar Osanlouy, Mona Khan, Soroush Safaei, Gonzalo Maso Talou, Maurice A Curtis, Peter Mombaerts
Publication: Communications Biology (November 2024)
Code: Zenodo Repository
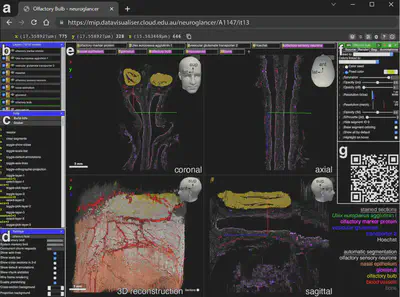
Interactive 3D Model: Neuroglancer Platform
Overview
This study presents a computational pipeline to reconstruct the 3D microanatomy of the human olfactory system, from the nasal cavity to the olfactory bulb—using fluorescence histology, deep learning, and high-performance computing (HPC). The workflow addresses challenges in processing terabyte-scale datasets and provides new insights into olfactory sensory neuron (OSN) distribution and axon trajectories.
Key Computational Contributions
- First end-to-end pipeline for large-scale 3D reconstruction of human olfactory tissues (~7.45 cm³ specimen, 1234 sections).
- CNN-based segmentation achieving Dice scores >0.85 for critical structures (OSNs, glomeruli, vasculature).
- HPC-optimized registration reducing banana-effect artifacts via multi-resolution deformable alignment.
- Public 3D dataset enabling interactive exploration of olfactory projection via Neuroglancer.
Methodology
Computational Pipeline

From fluorescence histology to 3D visualization – click to expand
-
Fluorescence Histology
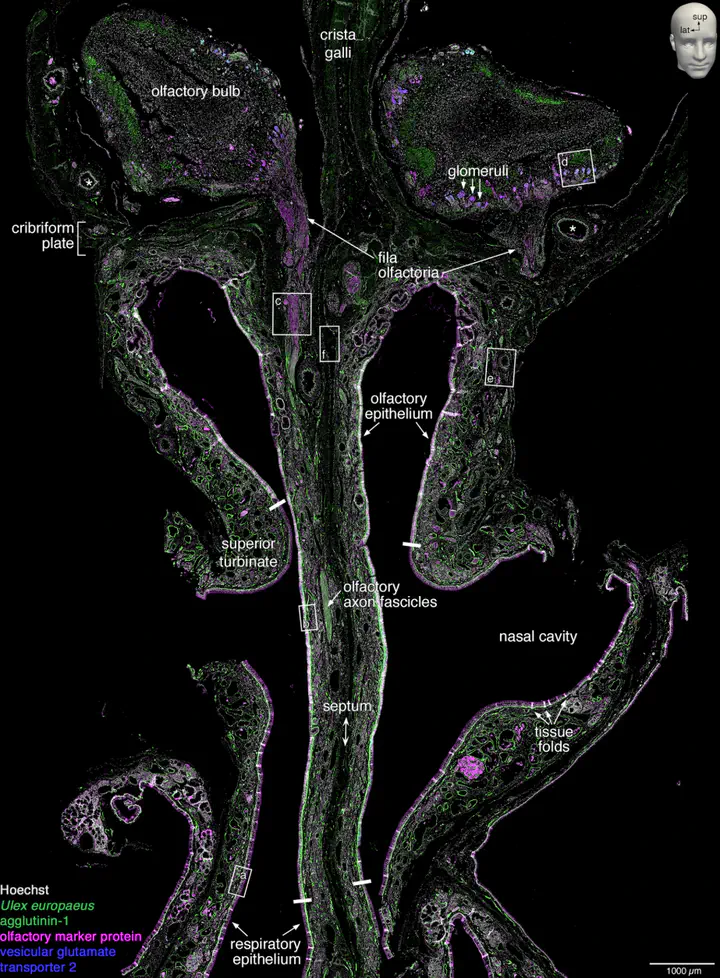
- Quadruple staining: Hoechst (nuclei), UEA1 (OSNs), OMP (mature OSNs), VGLUT2 (axon terminals).
- Whole-slide scanning: 1.097 µm/pixel resolution, ~2.9 TB raw data.
-
CNN Segmentation
- Architecture: Modified 2D U-Net with 4 input channels (Hoechst + 3 markers).
- Training: Bootstrap approach with iterative ground truth expansion (20-45 sections/structure).
- Key layers:
# Simplified U-Net structure encoder = [Conv2D(8→16→32), MaxPooling2D] bottleneck = Conv2D(64) + Upsampling2D decoder = [Conv2D(32→16→8), Concatenate(skip connections)]
-
HPC Registration
- Intra-block: Parallel registration of 247 blocks (5 sections each) using SimpleElastix.
- Inter-block: Banana-effect correction via affine + B-spline transformations between blocks.
- Metrics: Mutual information for intensity alignment, DSC for structural consistency.
Results
Segmentation Performance
| Structure | Dice Score | Binary Cross-Entropy |
|---|---|---|
| Vasculature | 0.808 | 0.0148 |
| OSNs | 0.760 | 0.0173 |
| Glomeruli | 0.779 | 0.0017 |
OSN segmentation achieved single-cell resolution in sparse regions but grouped cells in dense zones.
Registration Efficiency
- 1082 CPU hours on 96 Intel Xeon Gold 6136 cores
- 16% error reduction vs. sequential registration
- Tolerance: ±80 µm axial drift corrected
Key Findings
- OSN Count: ~2.7 million OSNs calculated via morphometric extrapolation (90% CI: 2.4–2.9M).
- Fila Olfactoria: 34 foramina identified in cribriform plate (17/side).
- Non-uniform Distribution: Olfactory epithelium showed serrated borders and posterior-anterior density gradient.
Implications
Technical Advancements
- Scalable ML: Method enables processing of whole-brain datasets (~100x mouse brain volume).
- Clinical Potential: Pipeline adaptable for Parkinson’s/Alzheimer’s studies via α-synuclein/tau staining.
- Open Science: First public 3D olfactory dataset with ~5.8 GB/channel resolution.
Biological Insights
- Challenges mouse-to-human extrapolation: ~10x fewer OSNs/glomerulus vs. mice.
- Provides baseline for studying SARS-CoV-2 olfactory dysfunction.
Computational Tools Used
- Segmentation: TensorFlow U-Net, Fiji for ground truth
- Registration: SimpleElastix, ITK
- Visualization: ParaView, Neuroglancer
- HPC: New Zealand eScience Infrastructure (NeSI)